Compound degraded:dibenzo-p-dioxin (DD)
General Description (About POP compound)
Polychlorinated dibenzodioxins (PCDDs), or simply dioxins, are a group of long-lived polyhalogenated organic compounds that are primarily anthropogenic, and contribute toxic, persistent organic pollution in the environment. Polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs) may be formed during the manufacture of chlorinated pesticides, and can remain in the products as impurities. There are 75 positional isomers of PCDDs
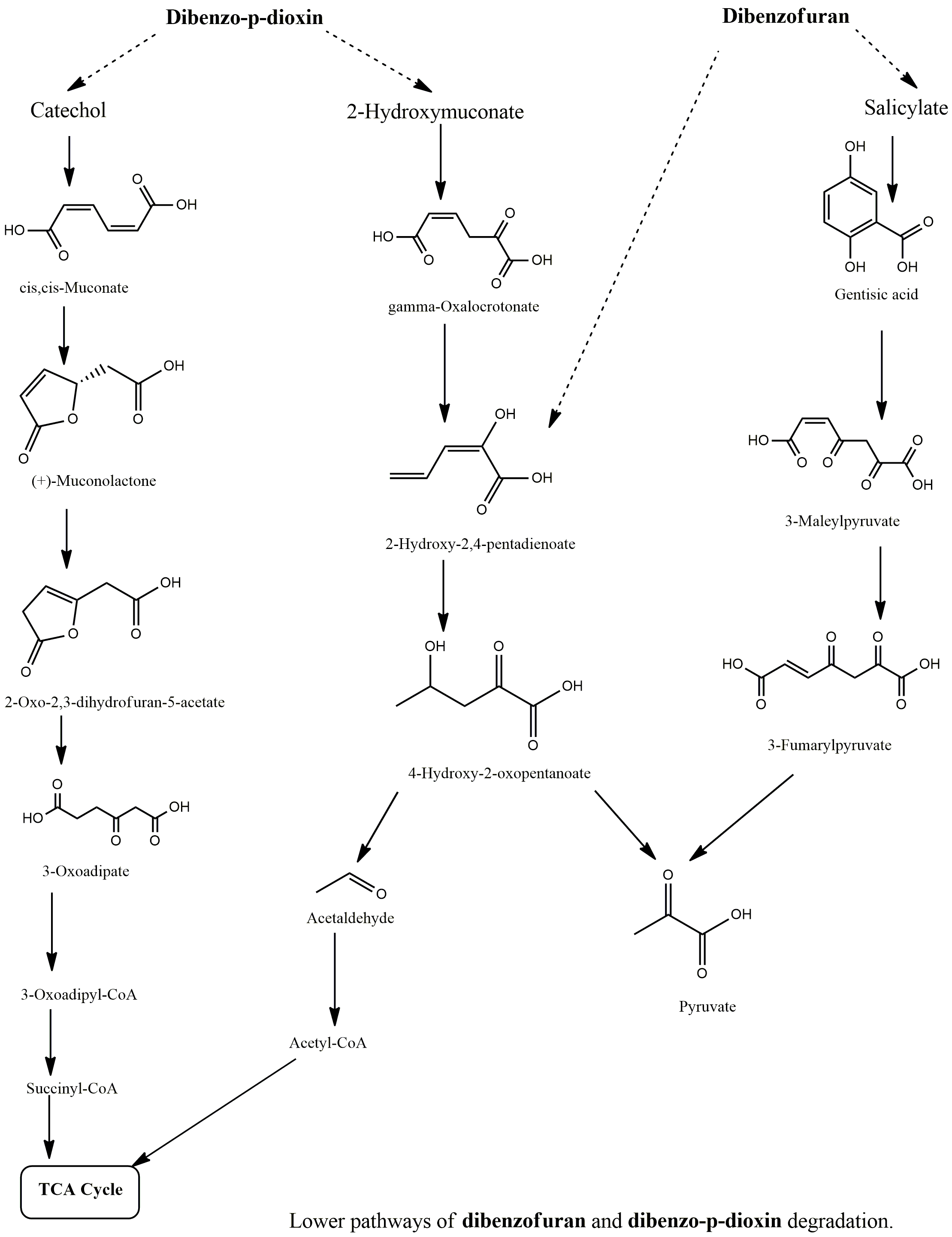
Biodegradation pathway
Publications
| Abstract | Title | Authors | Article Link |
|---|---|---|---|
| Sphingomonas wittichii strain RW1 (RW1) is one of the few strains that can grow on dibenzo-p-dioxin (DD). We conducted a transcriptomic study of RW1 using RNA-Seq to outline transcriptional responses to DD, dibenzofuran (DF), and the smectite clay mineral saponite with succinate as carbon source. The ability to grow on DD is rare compared to growth on the chemically similar DF even though the same initial dioxygenase may be involved in oxidation of both substrates. Therefore, we hypothesized the reason for this lies beyond catabolic pathways and may concern genes involved in processes for cell-substrate interactions such as substrate recognition, transport, and detoxification. Compared to succinate (SUC) as control carbon source, DF caused over 240 protein-coding genes to be differentially expressed, whereas more than 300 were differentially expressed with DD. Stress response genes were up-regulated in response to both DD and DF. This effect was stronger with DD than DF, suggesting a higher toxicity of DD compared to DF. Both DD and DF caused changes in expression of genes involved in active cross-membrane transport such as TonB-dependent receptor proteins, but the patterns of change differed between the two substrates. Multiple transcription factor genes also displayed expression patterns distinct to DD and DF growth. DD and DF induced the catechol ortho- and the salicylate/gentisate pathways, respectively. Both DD and DF induced the shared down-stream aliphatic intermediate compound pathway. Clay caused category-wide down-regulation of genes for cell motility and chemotaxis, particularly those involved in the synthesis, assembly and functioning of flagella. This is an environmentally important finding because clay is a major component of soil microbes’ microenvironment influencing local chemistry and may serve as a geosorbent for toxic pollutants. Similar to clay, DD and DF also affected expression of genes involved in motility and chemotaxis. | Sphingomonas wittichii Strain RW1 Genome-Wide Gene Expression Shifts in Response to Dioxins and Clay | Chai et al., 2016 | Link |
| Sphingomonas sp. strain RW1 synthesized a constitutive enzyme system that oxygenated dibenzofuran (DBF) to 2,2',3-trihydroxybiphenyl (THB). We purified this dibenzofuran 4,4a-dioxygenase system (DBFDOS) and found it to consist of four components which catalyzed three activities. Two isofunctional, monomeric flavoproteins (components A1 and A2; M(r) of about 44,000) transferred electrons from NADH to the second component (B; M(r) of about 12,000), a ferredoxin, which transported electrons to the heteromultimeric (alpha 2 beta 2) oxygenase component (C; M(r) of alpha, 45,000; M(r) of beta, 23,000). DBFDOS consumed 1 mol each of NADH, O2, and DBF, which was dioxygenated to about 1 mol of THB; no intermediate was observed. The reaction was thus the dioxygenation of DBF at the 4 and 4a positions to give a diene-diol-hemiacetal which rearomatized by spontaneous loss of a phenolate group to form THB. Components A1 and A2 each reduced dichlorophenolindophenol but had negligible activity with cytochrome c; each lost the yellow color, observed to be flavin adenine dinucleotide, upon purification. Component B, which transported electrons to the oxygenase or cytochrome c, had an N-terminal amino acid sequence with high homology to the putidaredoxin of cytochrome P-450cam. The oxygenase had the UV spectrum of a Rieske iron-sulfur center. We presume DBFDOS to be a class IIA dioxygenase system (EC 1.14.12.-), functionally similar to pyrazon dioxygenase. | Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system | Bünz and Cook. 1993 | Link |
| A gram-positive bacterium Terrabacter sp. strain DBF63 is able to degrade dibenzofuran (DF) via initial dioxygenation by a novel angular dioxygenase. The dbfA1 and dbfA2 genes, which encode the large and small subunits of the dibenzofuran 4,4a-dioxygenase (DFDO), respectively, were isolated by a polymerase chain reaction-based method. DbfA1 and DbfA2 showed moderate homology to the large and small subunits of other ring-hydroxylating dioxygenases (less than 40%), respectively, and some motifs such as the Fe(II) binding site and the [2Fe-2S] cluster ligands were conserved in DbfA1. DFDO activity was confirmed in Escherichia coli cells containing the cloned dbfA1 and dbfA2 genes with the complementation of nonspecific ferredoxin and ferredoxin reductase component of E. coli. Under this condition, these cells exhibited angular dioxygenation of DF and dibenzo-p-dioxin, and monooxygenation of fluorene, but not angular dioxygenation of carbazole, xanthene, and phenoxathiin. Phylogenetic analysis revealed that DbfA1 formed a branch with recently reported large subunits of polycyclic aromatic hydrocarbon (PAH) dioxygenase from gram-positive bacteria but did not cluster with that of other angular dioxygenases, i.e., DxnA1 from Sphingomonas sp. strain RW1 [Armengaud, J., Happe, B., and Timmis, K. N. J. Bacteriol. 180, 3954-3966, 1998] and CarAa from Pseudomonas sp. strain CA10 [Sato, S., Nam, J.-W., Kasuga, K., Nojiri, H., Yamane, H., and Omori, T. J. Bacteriol. 179, 4850-4858, 1997]. | Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63 | Kasuga et al., 2001 | Link |
| Spore-forming bacterial strains capable of utilizing dibenzofuran (DF) as a sole source of carbon and energy were isolated. Characteristics of the isolates justified their classification into the genus Paenibacillus, and their closest relative was P. naphthalenovorans. Degenerate primers for aromatic hydrocarbon dioxygenase alpha subunit (AhDOa) genes and genomic DNA of the strain YK5 were used for gene isolation. The nucleotide sequences of clones of the PCR products revealed that the strain YK5 carries at least five different AhDOa genes. Northern hybridization analysis showed that one of the AhDOa genes was transcribed under DF-containing culture conditions. A gene cluster encoding the AhDOa was isolated. The genes predicted to encode extradiol dioxygenase (dbfB) and hydrolase (dbfC) were found to be an upstream of genes encoding the alpha and beta subunit of the AhDO (dbfA1 and dbfA2, respectively); the latter two gene products showed 60 and 53% identity to the amino acid sequences of DbfA1 and DbfA2 of Terrabacter sp. DBF63, respectively. Two Paenibacillus validus JCM 9077 strains transformed with the dbf gene clusters acquired the ability to convert DF to 2,2?,3-trihydroxybiphenyl (THBP) and salicylic acid (SAL). These results suggest that the enzymes encoded by the gene cluster isolated in this study are involved in DF metabolism in YK5. | Isolation and characterization of a gene cluster for dibenzofuran degradation in a new dibenzofuran-utilizing bacterium, Paenibacillus sp. strain YK5 | Lida et al., 2005 | Link |
| A key enzyme in the degradation pathways of dibenzo-p-dioxin and dibenzofuran, namely, 2,2',3-trihydroxybiphenyl dioxygenase, which is responsible for meta cleavage of the first aromatic ring, has been genetically and biochemically analyzed. The dbfB gene of this enzyme has been cloned from a cosmid library of the dibenzo-p-dioxin- and dibenzofuran-degrading bacterium Sphingomonas sp. strain RW1 (R. M. Wittich, H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel, Appl. Environ. Microbiol. 58:1005-1010, 1992) and sequenced. The amino acid sequence of this enzyme is typical of those of extradiol dioxygenases. This enzyme, which is extremely oxygen labile, was purified anaerobically to apparent homogeneity from an Escherichia coli strain that had been engineered to hyperexpress dbfB. Unlike most extradiol dioxygenases, which have an oligomeric quaternary structure, the 2,2',3-trihydroxybiphenyl dioxygenase is a monomeric protein. Kinetic measurements with the purified enzyme produced similar Km values for 2,2',3-trihydroxybiphenyl and 2,3-dihydroxybiphenyl, and both of these compounds exhibited strong substrate inhibition. 2,2',3-Trihydroxydiphenyl ether, catechol, 3-methylcatechol, and 4-methylcatechol were oxidized less efficiently and 3,4-dihydroxybiphenyl was oxidized considerably less efficiently. | Characterization of 2,2',3-trihydroxybiphenyl dioxygenase, an extradiol dioxygenase from the dibenzofuran- and dibenzo-p-dioxin-degrading bacterium Sphingomonas sp. strain RW1. | Happe et al., 1993 | Link |
| Sphingomonas wittichii RW1 is one of a few strains known to grow on the related compounds dibenzofuran (DBF) and dibenzo-p-dioxin (DXN) as the sole source of carbon. Previous work by others (B. Happe, L. D. Eltis, H. Poth, R. Hedderich, and K. N. Timmis, J Bacteriol 175:7313–7320, 1993, https://doi.org/10.1128/jb.175.22.7313-7320.1993) showed that purified DbfB had significant ring cleavage activity against the DBF metabolite trihydroxybiphenyl but little activity against the DXN metabolite trihydroxybiphenylether. We took a physiological approach to positively identify ring cleavage enzymes involved in the DBF and DXN pathways. Knockout of dbfB on the RW1 megaplasmid pSWIT02 results in a strain that grows slowly on DBF but normally on DXN, confirming that DbfB is not involved in DXN degradation. Knockout of SWIT3046 on the RW1 chromosome results in a strain that grows normally on DBF but that does not grow on DXN, demonstrating that SWIT3046 is required for DXN degradation. A double-knockout strain does not grow on either DBF or DXN, demonstrating that these are the only ring cleavage enzymes involved in RW1 DBF and DXN degradation. The replacement of dbfB by SWIT3046 results in a strain that grows normally (equal to the wild type) on both DBF and DXN, showing that promoter strength is important for SWIT3046 to take the place of DbfB in DBF degradation. Thus, both dbfB- and SWIT3046-encoded enzymes are involved in DBF degradation, but only the SWIT3046-encoded enzyme is involved in DXN degradation. | Separate Upper Pathway Ring Cleavage Dioxygenases Are Required for Growth of Sphingomonas wittichii Strain RW1 on Dibenzofuran and Dibenzo- p-Dioxin | Mutter and Zylstra. 2021 | Link |
| Sphingomonas wittichii RW1 grows on the two related compounds dibenzofuran (DBF) and dibenzo-p-dioxin (DXN) as the sole source of carbon. Previous work by others (P. V. Bunz, R. Falchetto, and A. M. Cook, Biodegradation 4:171-178, 1993, https://doi/org/10.1007/BF00695119) identified two upper pathway meta cleavage product hydrolases (DxnB1 and DxnB2) active on the DBF upper pathway metabolite 2-hydroxy-6-oxo-6-(2-hydroxyphenyl)-hexa-2,4-dienoate. We took a physiological approach to determine the role of these two enzymes in the degradation of DBF and DXN by RW1. Single knockouts of either plasmid-located dxnB1 or chromosome-located dxnB2 had no effect on RW1 growth on either DBF or DXN. However, a double-knockout strain lost the ability to grow on DBF but still grew normally on DXN, demonstrating that DxnB1 and DxnB2 are the only hydrolases involved in the DBF upper pathway. Using a transcriptomics-guided approach, we identified a constitutively expressed third hydrolase encoded by the chromosomally located SWIT0910 gene. Knockout of SWIT0910 resulted in a strain that no longer grows on DXN but still grows normally on DBF. Thus, the DxnB1 and DxnB2 hydrolases function in the DBF but not the DXN catabolic pathway, and the SWIT0190 hydrolase functions in the DXN but not the DBF catabolic pathway. IMPORTANCE S. wittichii RW1 is one of only a few strains known to grow on DXN as the sole source of carbon. Much of the work deciphering the related RW1 DXN and DBF catabolic pathways has involved genome gazing, transcriptomics, proteomics, heterologous expression, and enzyme purification and characterization. Very little research has utilized physiological techniques to precisely dissect the genes and enzymes involved in DBF and DXN degradation. Previous work by others identified and extensively characterized two RW1 upper pathway hydrolases. Our present work demonstrates that these two enzymes are involved in DBF but not DXN degradation. In addition, our work identified a third constitutively expressed hydrolase that is involved in DXN but not DBF degradation. Combined with our previous work (T. Y. Mutter and G. J. Zylstra, Appl Environ Microbiol 87:e02464-20, 2021, https://doi.org/10.1128/AEM.02464-20), this means that the RW1 DXN upper pathway involves genes from three very different locations in the genome, including an initial plasmid-encoded dioxygenase and a ring cleavage enzyme and hydrolase encoded on opposite sides of the chromosome. | Differential Roles of Three Different Upper Pathway meta Ring Cleavage Product Hydrolases in the Degradation of Dibenzo- p-Dioxin and Dibenzofuran by Sphingomonas wittichii Strain RW1 | Mutter and Zylstra. 2021 | Link |
| In the course of our screening for dibenzo-p-dioxin-utilizing bacteria, a Sphingomonas sp. strain was isolated from enrichment cultures inoculated with water samples from the river Elbe. The isolate grew with both the biaryl ethers dibenzo-p-dioxin and dibenzofuran (DF) as the sole sources of carbon and energy, showing doubling times of about 8 and 5 h, respectively. Biodegradation of the two aromatic compounds initially proceeded after an oxygenolytic attack at the angular position adjacent to the ether bridge, producing 2,2',3-trihydroxydiphenyl ether or 2,2',3-trihydroxybiphenyl from the initially formed dihydrodiols, which represent extremely unstable hemiacetals. Results obtained from determinations of enzyme activities and oxygen consumption suggest meta cleavage of the trihydroxy compounds. During dibenzofuran degradation, hydrolysis of 2-hydroxy-6-oxo-6-(2-hydroxyphenyl)-hexa-2,4-dienoate yielded salicylate, which was branched into the catechol meta cleavage pathway and the gentisate pathway. Catechol obtained from the product of meta ring fission of 2,2',3-trihydroxydiphenyl ether was both ortho and meta cleaved by Sphingomonas sp. strain RW1 when this organism was grown with dibenzo-p-dioxin. | Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1 | Wittich et al., 1992 | Link |
| Sphingomonas wittichii RW1 is a bacterium of interest due to its ability to degrade polychlorinated dioxins, which represent priority pollutants in the USA and worldwide. Although its genome has been fully sequenced, many questions exist regarding changes in protein expression of S. wittichii RW1 in response to dioxin metabolism. We used difference gel electrophoresis (DIGE) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) to identify proteomic changes induced by growth on dibenzofuran, a surrogate for dioxin, as compared to acetate. Approximately 10% of the entire putative proteome of RW1 could be observed. Several components of the dioxin and dibenzofuran degradation pathway were shown to be upregulated, thereby highlighting the utility of using proteomic analyses for studying bioremediation agents. This is the first global protein analysis of a microorganism capable of utilizing the carbon backbone of both polychlorinated dioxins and dibenzofurans as the sole source for carbon and energy. | Proteomic Profiling of the Dioxin-Degrading Bacterium Sphingomonas wittichii RW1 | Colquhoun et al., 2012 | Link |
| Aerobic biotransformation of the diaryl ethers 2,7-dichlorodibenzo-p-dioxin and 1,2,3,4-tetrachlorodibenzo-p-dioxin by the dibenzo-p-dioxin-utilizing strain Sphingomonas wittichii RW1, producing corresponding metabolites, was demonstrated for the first time. Our strain transformed 2,7-dichlorodibenzo-p-dioxin, yielding 4-chlorocatechol, and 1,2,3,4-tetrachlorodibenzo-p-dioxin, producing 3,4,5,6-tetrachlorocatechol and 2-methoxy-3,4,5,6-tetrachlorophenol; all of these compounds were unequivocally identified by mass spectrometry both before and after N,O-bis(trimethylsilyl)-trifluoroacetamide derivatization by comparison with authentic standards. Additional experiments showed that strain RW1 formed a second metabolite, 2-methoxy-3,4,5,6-tetrachlorophenol, from the original degradation product, 3,4,5,6-tetrachlorocatechol, by methylation of one of the two hydroxy substituents. | Biotransformation of 2,7-Dichloro- and 1,2,3,4-Tetrachlorodibenzo-p-Dioxin by Sphingomonas wittichii RW1 | Hong et al., 2002 | Link |
| Density functional theory (DFT) calculations were used to explore the relationship between the biotransformation of dibenzo-p-dioxin and selected chlorinated derivatives by resting cells of Sphingomonas wittichii RW1 and measuring the thermodynamic properties of the biotransformation substrates. Sphingomonas wittichii RW1 can aerobically catabolize dibenzo-p-dioxin as well as 2,7-dichloro-, 1,2,3-trichloro-, 1,2,3,4-tetrachloro-, and 1,2,3,4,7,8-hexachlorodibenzo-p-dioxin; however, neither the 2,3,7-trichloro- nor the 1,2,3,7,8-pentachlorodibenzo-p-dioxin was transformed to its corresponding metabolic intermediate. The experimental biotransformation rates established were apparently governed by the selected thermodynamic properties of the substrates tested. | Is the biotransformation of chlorinated dibenzo-p-dioxins by Sphingomonas wittichii RW1 governed by thermodynamic factors? | Nam et al., 2014 | Link |
| Anaerobic enrichment cultures derived from contaminated Kymijoki River sediments dechlorinated 1,2,3,4-tetrachlorodibenzofuran (1,2,3,4-tetra-CDF), octachlorodibenzofuran (octa-CDF) and 1,2,3,4-tetrachlorodibenzo-p-dioxin (1,2,3,4-tetra-CDD). 1,2,3,4-tetra-CDF was dechlorinated via 1,2,3-, 2,3,4-, and 1,3,4/1,2,4-tri-CDFs to 1,3-, 2,3-, and 2,4-di-CDFs and finally to 4-mono-CDF. The dechlorination rate of 1,2,3,4-tetra-CDF was generally slower than that of 1,2,3,4-tetra-CDD. The rate and extent of 1,2,3,4-tetra-CDD dechlorination was enhanced by addition of pentachloronitrobenzene (PCNB) as a co-substrate. Dechlorination of spiked octa-CDF was observed with the production of hepta-, hexa-, penta- and tetra-CDFs over 6 months. Two major phylotypes of the Chloroflexi community showed an increase, one of which was identical to the Dehalococcoides mccartyi Pinellas subgroup. A set of twelve putative reductive dehalogenase (rdh) genes increased in abundance with addition of 1,2,3,4-tetra-CDF, 1,2,3,4-tetra-CDD and/or PCNB. This information will aid in understanding how indigenous microbial communities impact the fate of PCDFs and in developing strategies for bioremediation of PCDD/F contaminated sediments. | Enriching for microbial reductive dechlorination of polychlorinated dibenzo-p-dioxins and dibenzofurans | Liu et al., 2014 | Link |
